Welcome to the documentation of the R package safuncs!
This package is designed to assist you in the analysis of your
biological data (e.g. survival).
Below I wrote a short tutorial on how some of the core functions in
safuncs can be used.
To get started, please install and load the package by running the following code:
remotes::install_github("sean4andrew/safuncs") # install package
library(safuncs) # loadSurvival Analysis
At ONDA, survival data is often available to scientists initially in
the form of rows of entries for every mortality observed. This data
needs to be converted to a useful form for analysis in R or Prism. The
function Surv_Gen() shortens the data transformation step
to just one line of code! Below, I showcase its use.
Let us first examine the raw survival data we can get from our OneDrive mort excel file:
data(mort_db_ex) # load example data file
head(mort_db_ex, n = 5) # view first 5 rows of data
#> Tank.ID Trt.ID TTE Status
#> 1 C1 B 0 1
#> 2 C6 B 4 1
#> 3 C6 B 7 1
#> 4 C1 B 19 1
#> 5 C8 D 19 1The 2nd-rightmost column named TTE indicates the Time to
Event, e.g. days post challenge, and the last column Status
indicates the type of event (1 = death, 0 = sampled-out or survived).
Missing from such a dataset are the rows representing surviving fish
(Status = 0); these needs to be present for a proper survival analysis.
Introducing Surv_Gen()!
Obtain the proper survival data by simply inserting the mort database
into Surv_Gen(). Provide a time (TTE) indicating until when
other fish survived and the starting number of fish used per tank.
Surv_Gen(mort_db = mort_db_ex,
starting_fish_count = 100, # per tank
last_tte = 54)Below is the output data’s bottom 5 rows which represent the generated survivors (Status = 0).
#> Tank.ID Trt.ID TTE Status
#> 1196 C8 D 54 0
#> 1197 C8 D 54 0
#> 1198 C8 D 54 0
#> 1199 C8 D 54 0
#> 1200 C8 D 54 0You also get some printout messages for verification purposes:
#> [1] "Your total number of tanks is: 12"
#> [1] "Your total number of treatment groups is: 4"
#> [1] "Your total number of fish in the output data is: 1200"You can also specify tank-specific starting fish numbers using a
dataframe as input to starting_fish_count!
# reduce example data to simplify the demonstration
mort_db_ex2 = mort_db_ex[mort_db_ex$Tank.ID %in% c("C01", "C02", "C03"),]
# apply Surv_Gen()!
Surv_Gen(mort_db = mort_db_ex2,
starting_fish_count = data.frame(Trt.ID = c("B", "A", "C"), # a vector of treatment groups for the specified tanks
Tank.ID = c("C1", "C2", "C3"), # a vector with ALL tanks in the study
starting_fish_count = c(100, 100, 50)), # a vector of fish numbers in same order as Tank.IDs
last_tte = 54)With a short R script as shown, you can quickly regenerate survival data whenever there is a slight change in the mort database; perhaps from day-to-day or after corrections.
For those wanting to use the generated survival data in prism, set
the argument output_prism = TRUE in Surv_Gen()
to save a prism-ready csv. in your working directory. If you want
starting and ending dates in the Prism output, input a starting date
into the argument output_prism_date, as shown:
Surv_Gen(mort_db = mort_db_ex,
starting_fish_count = 100, # per tank
last_tte = 54,
output_prism = TRUE,
output_prism_date = "08-Aug-2024")Once the complete survival data has been obtained, plots can be
easily generated using Surv_Plots()!
data(surv_db_ex) # load example complete survival data
Surv_Plots(surv_db_ex, # plot the survival curve and hazard curve
dailybin = FALSE) # recommended argument for good (large) sample size datasets
#> $Survival_Plot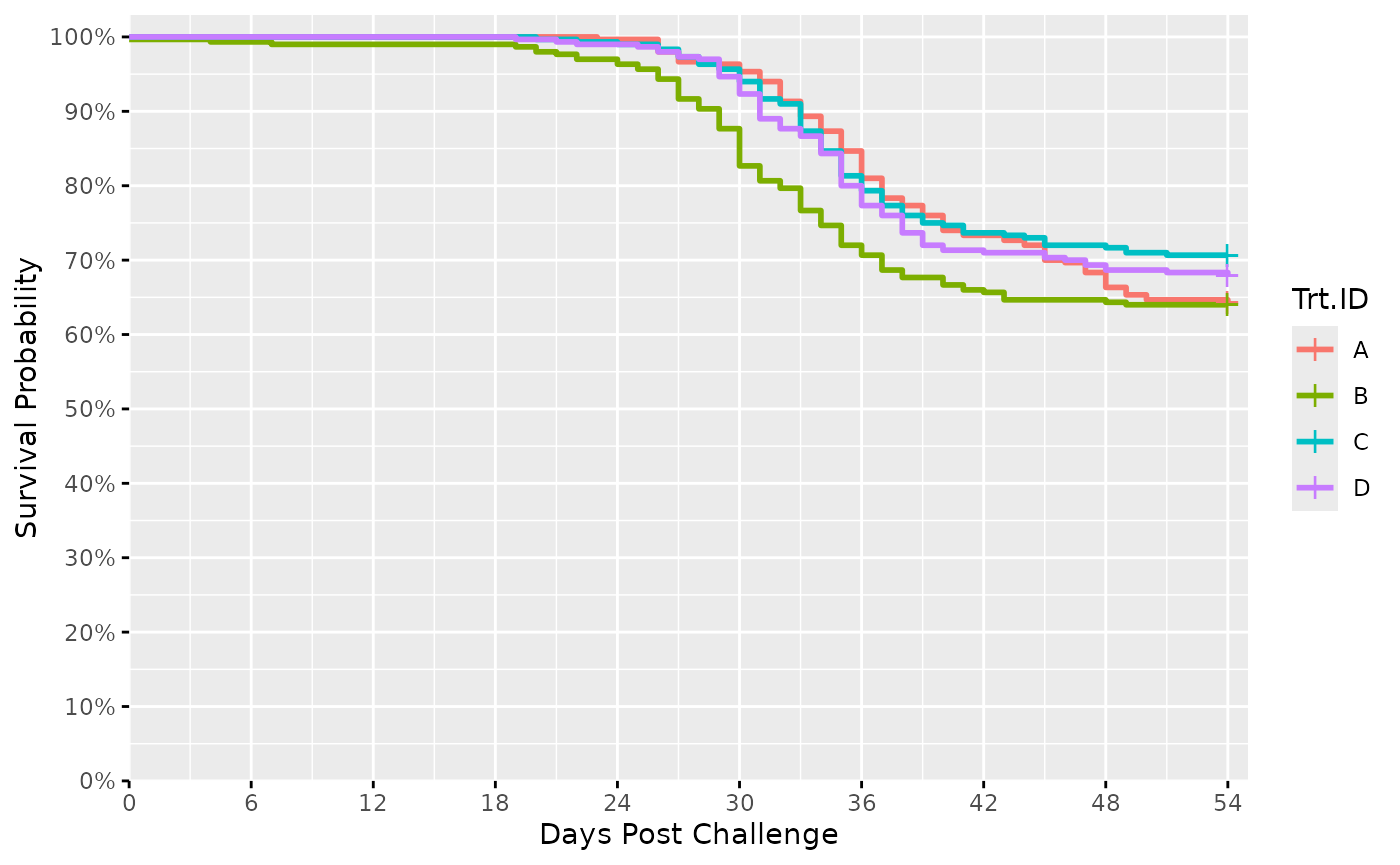
#>
#> $Hazard_Plot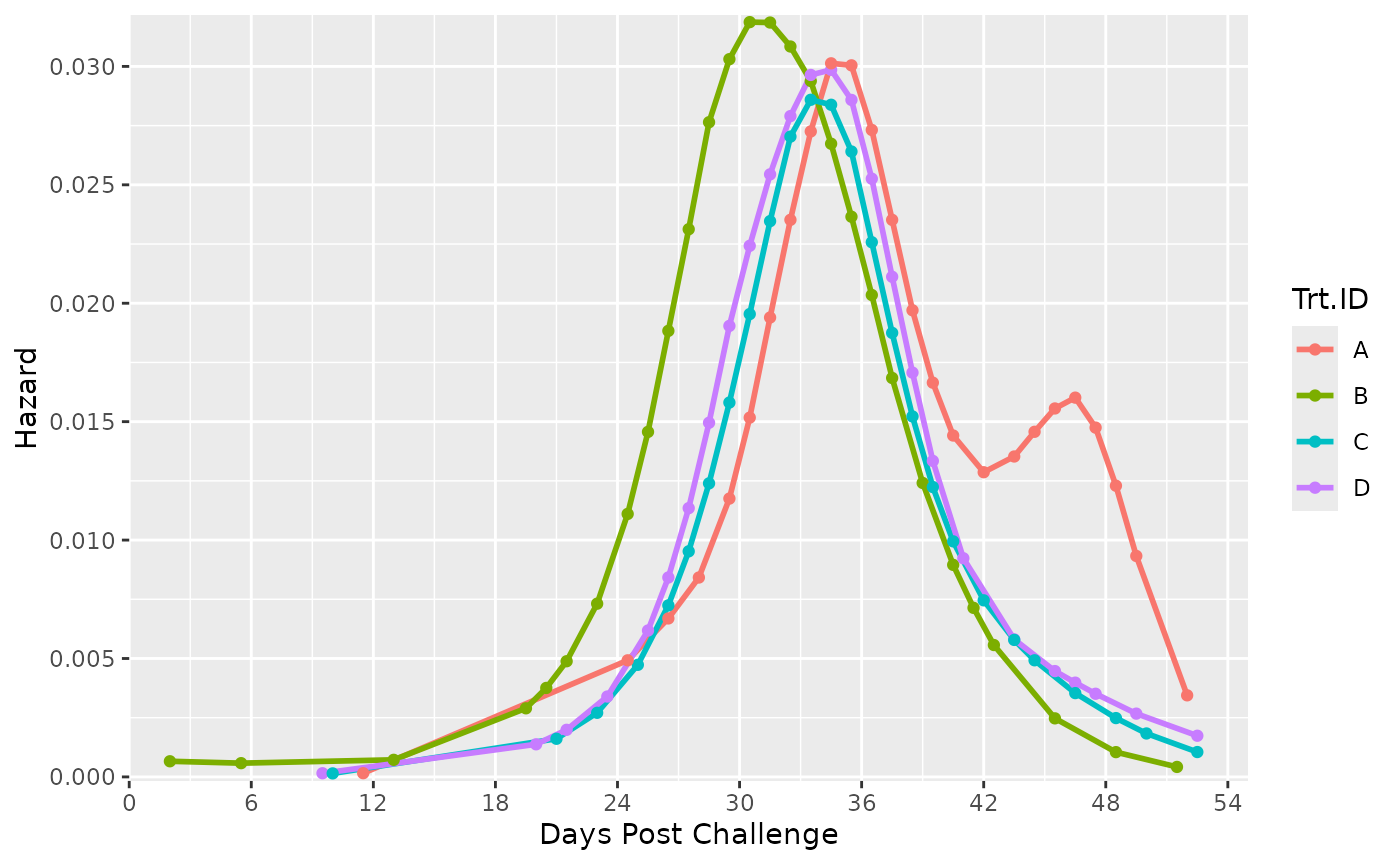 The plots save automatically to your working directory as a .tiff and in
a power-point editable format (.pptx). Notably,
The plots save automatically to your working directory as a .tiff and in
a power-point editable format (.pptx). Notably,
Surv_Plots() accepts many more arguments that can influence
the hazard plot in particular and needs tailoring for your specific
experiment. For more details on these arguments, please see the
functions’ documentation
page.
############TO BE CONTINUED###########
